Explaining ventilators for COVID-19
April 1 2020 · CoVent-19
Last week I collaborated with Dr. Sarah Low to explain ventilators in a series of animations. Sarah is an anesthesiology resident at Massachusetts General Hospital, and a leader of CoVent-19 - one of many efforts to address the ventilator shortage during the Covid-19 outbreak. The CoVent-19 organization is run by a team of thirteen medical doctors, including Sarah. Their goal is to teach engineers the basics of medical ventilation, in the hopes that new ventilation solutions can be engineered from existing equipment or repurposed supplies.
Please take a look at the CoVent-19 website for more information! I’m also sharing all of these ventilation graphics to the public domain, so you’re free to use them however you like . But please note that I only illustrated the simplified circuits that Sarah needed (this isn’t a comprehensive explanation of ventilators).
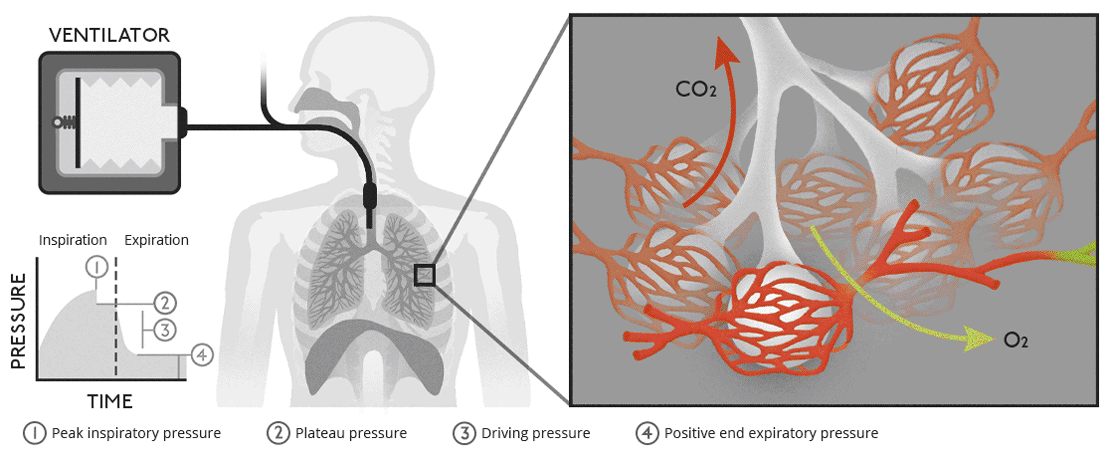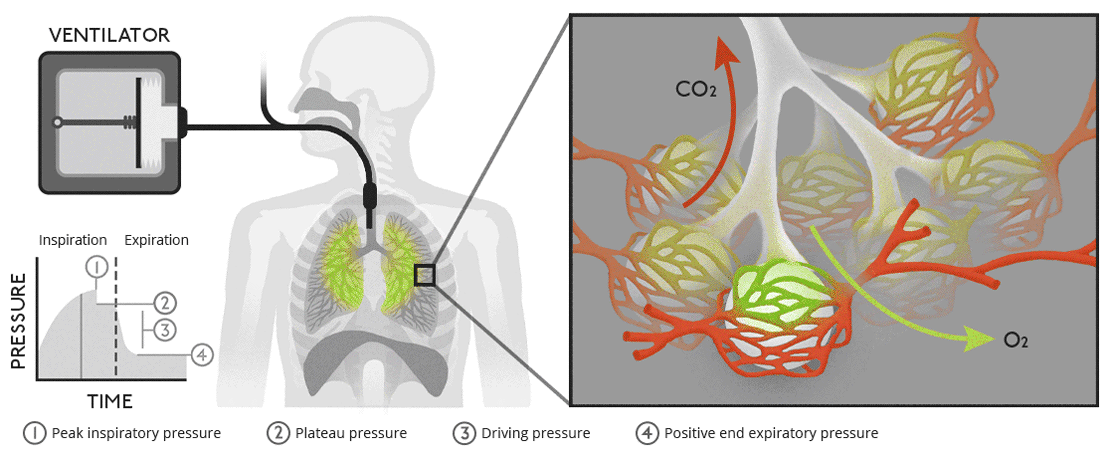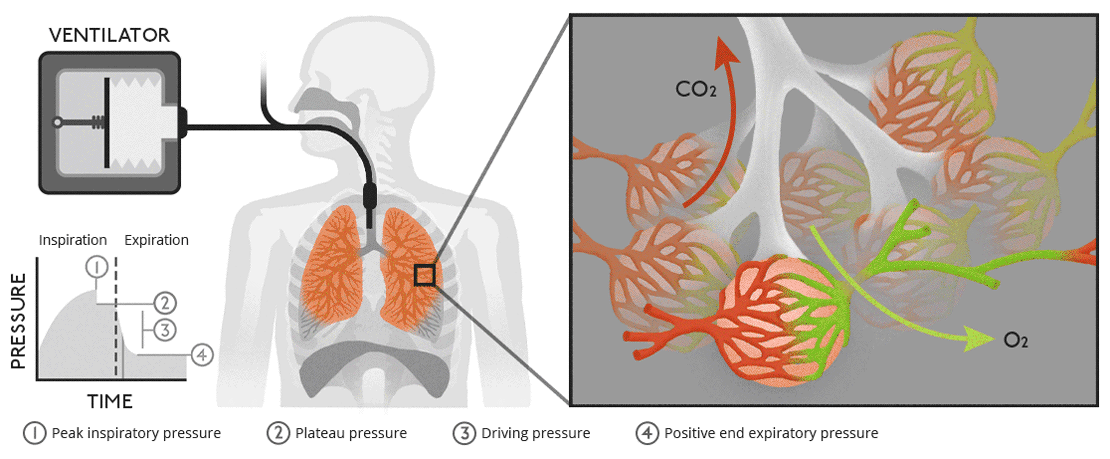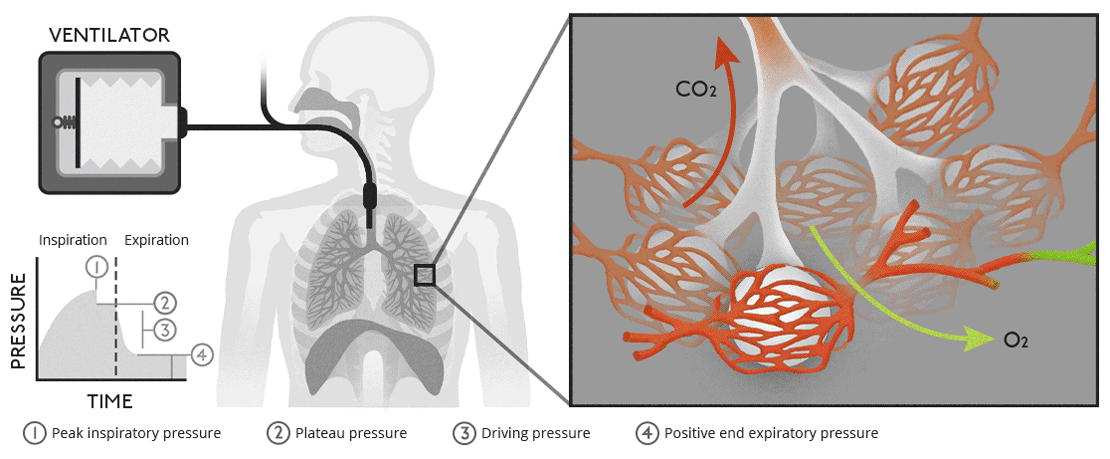
Biology of ventilationAir moves from our lungs into the bloodstream through tiny, air-filled sacs called alveoli. |
|
Open circuit ventilatorsThis is a very simple version of a ventilator where waste gas is expelled from the system without recycling. |
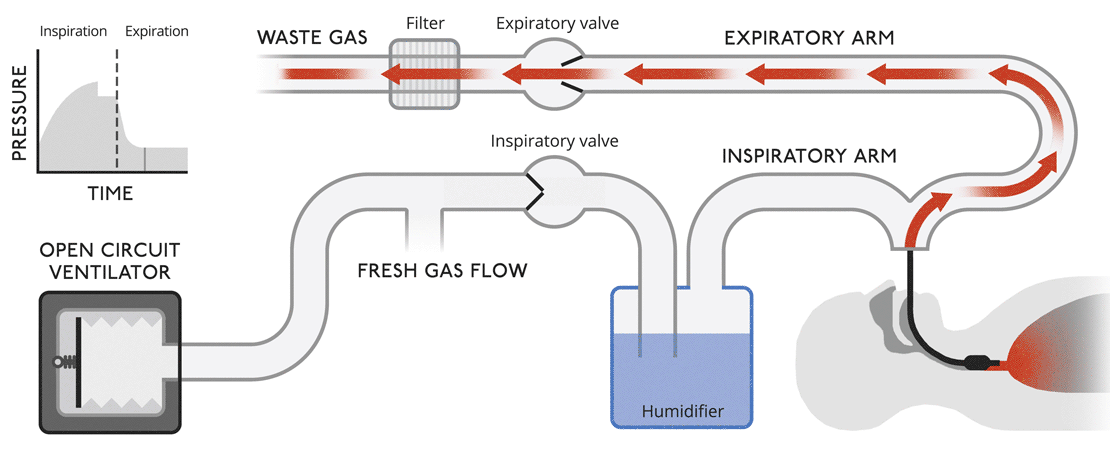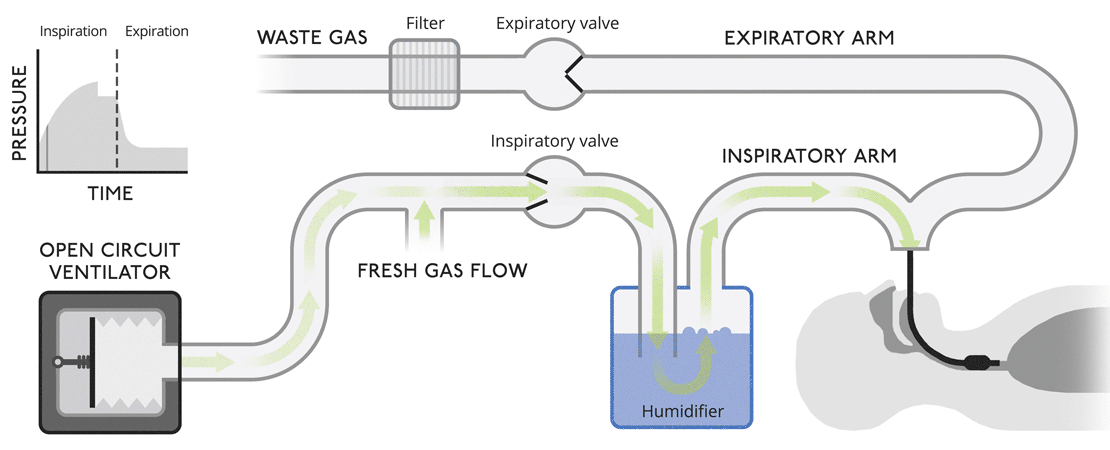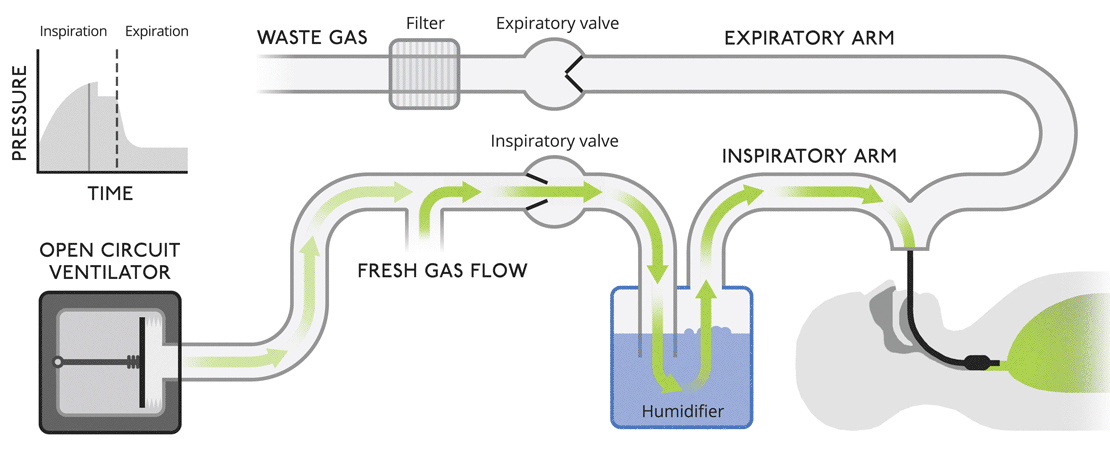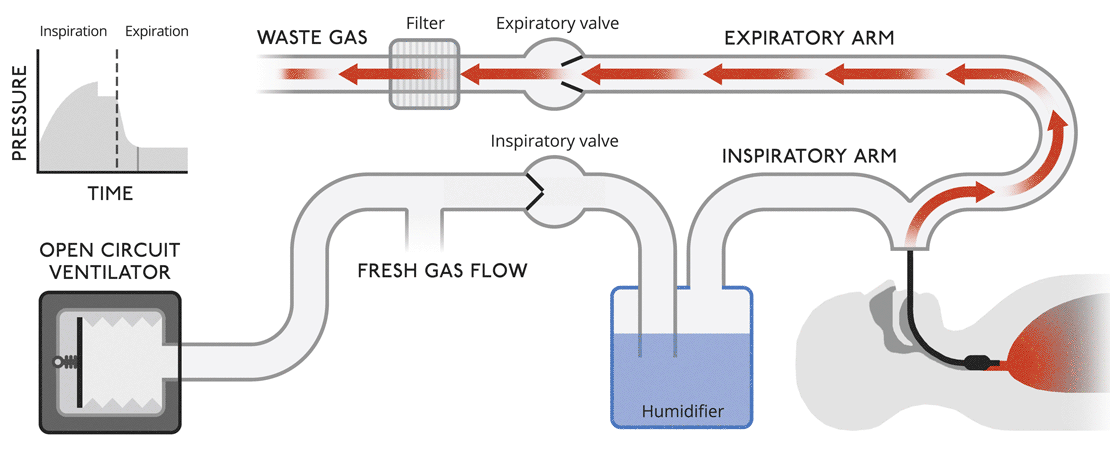
|
Closed circuit ventilatorsThis diagram is more like a modern medical ventilator. A CO |
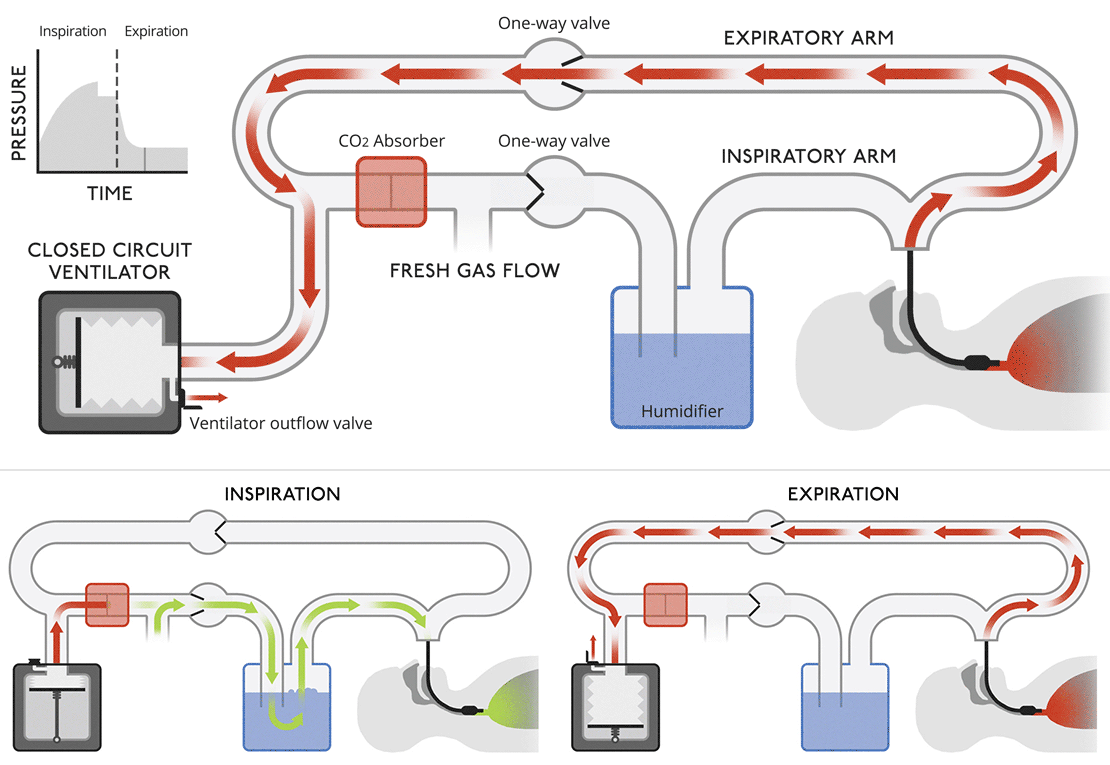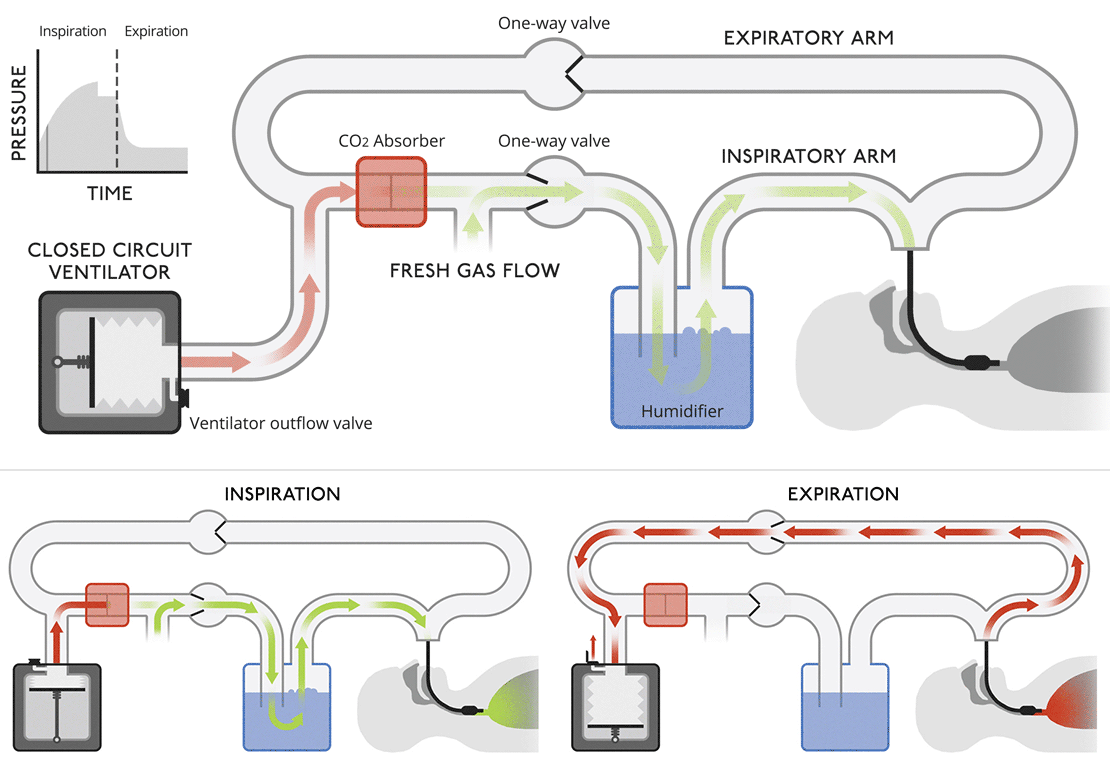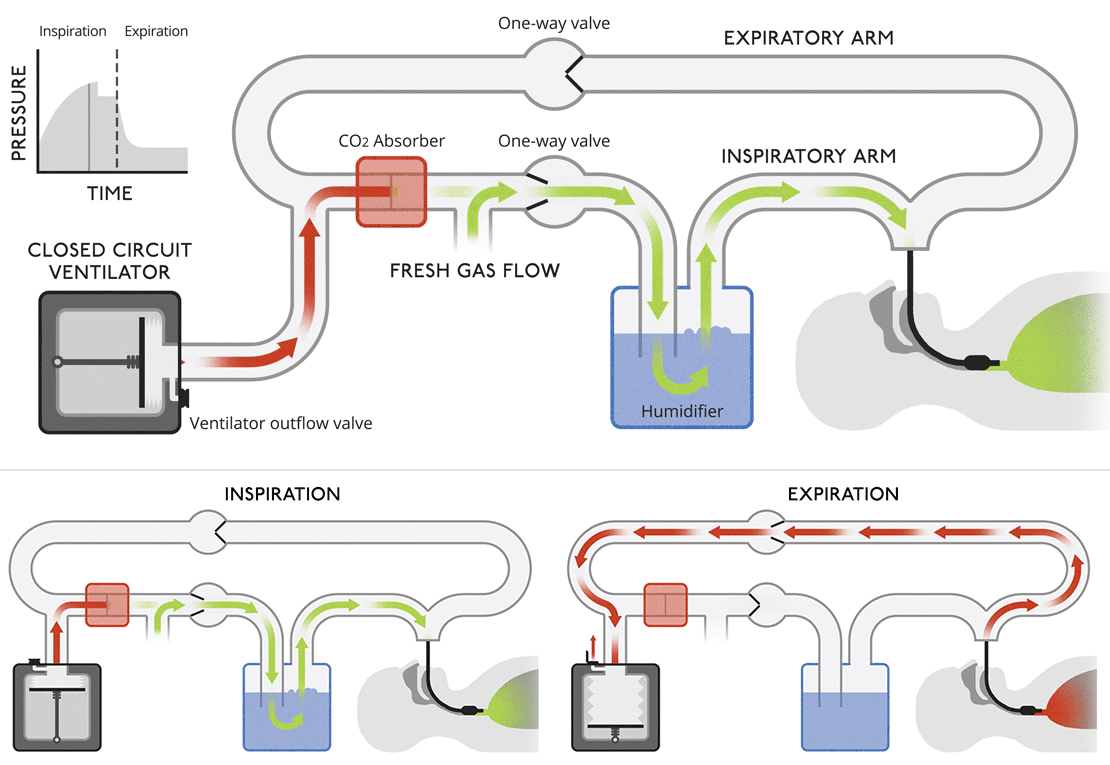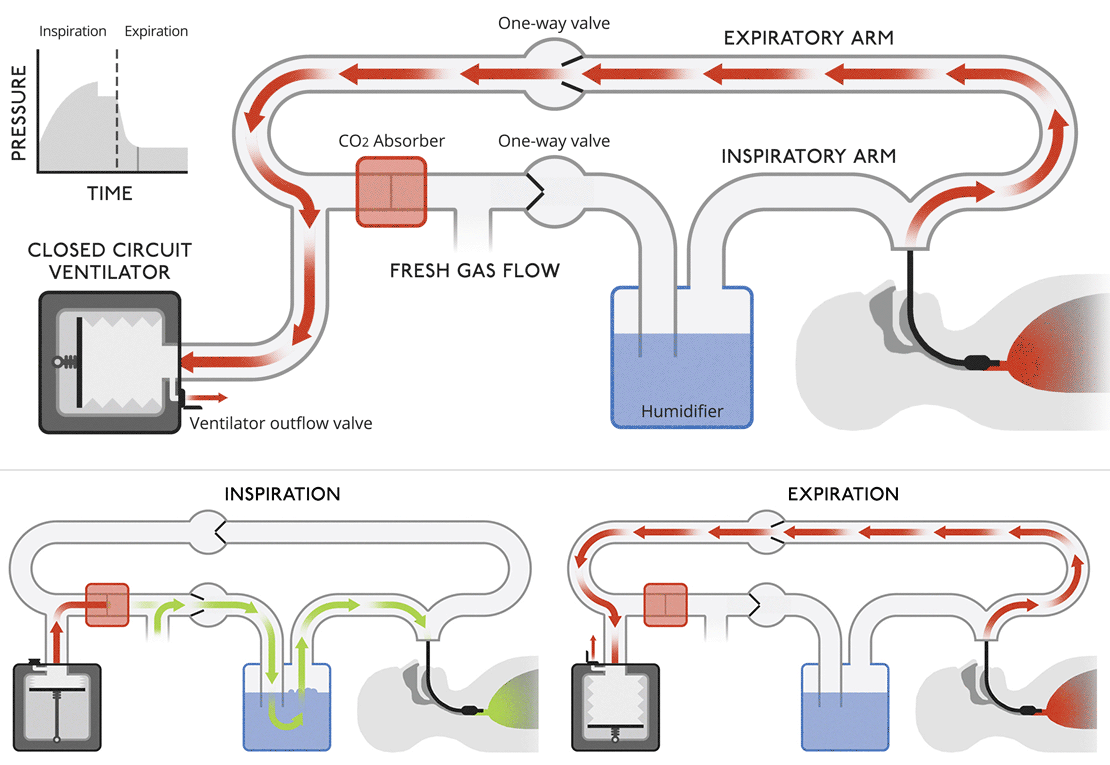
|
Biology of ventilationAir moves from our lungs into the bloodstream through tiny, air-filled sacs called alveoli. 
|
Open circuit ventilatorsThis is a very simple version of a ventilator where waste gas is expelled from the system without recycling. 
|
Closed circuit ventilatorsThis diagram is more like a modern medical ventilator. A CO 
|
The Geology of the Moon
August 26 2019 · Link to the Open-Source Code
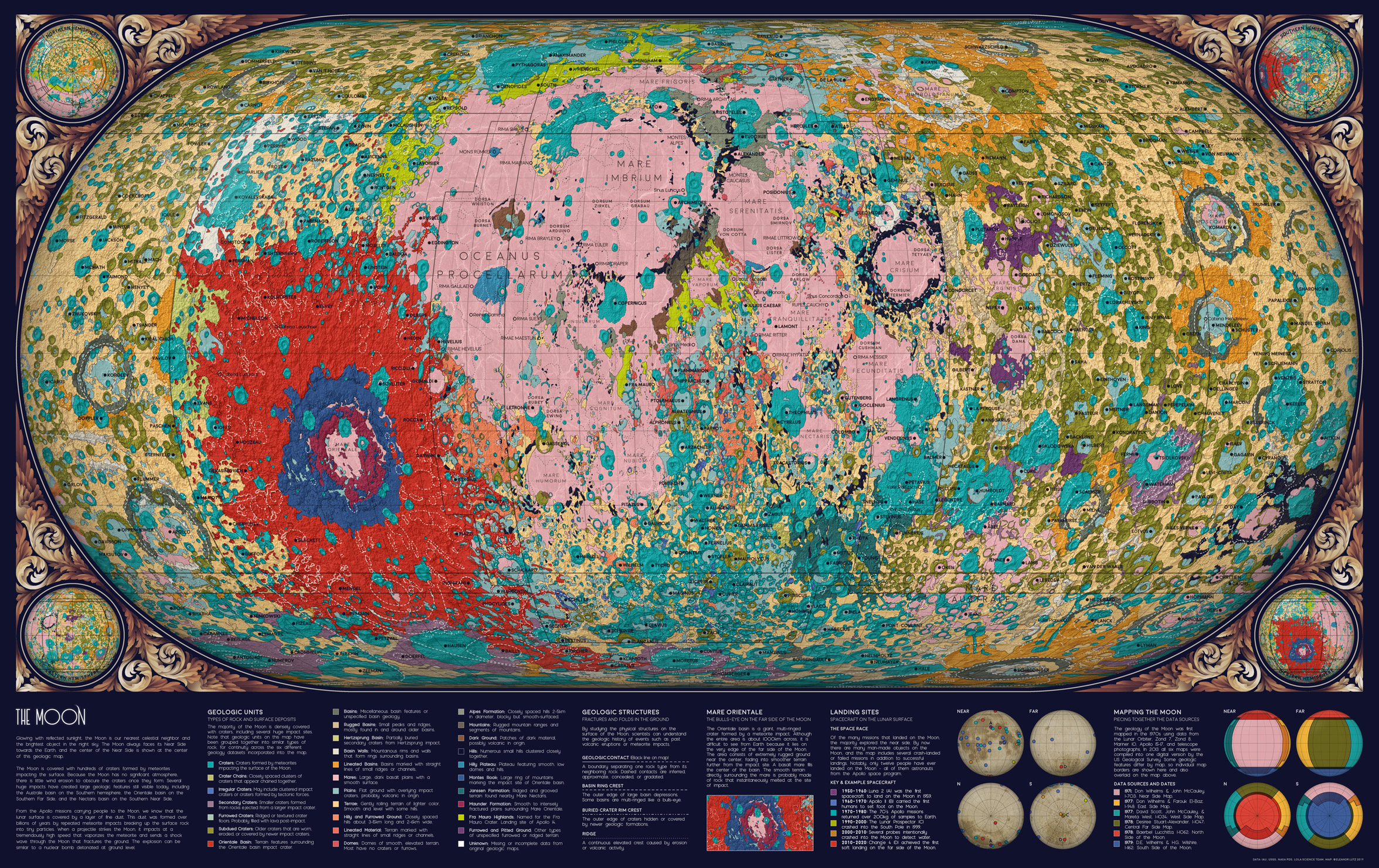
This map was one of my favorites, so I’m excited to share the code tutorial for a geologic map of the Moon! Although I already made a geologic map of Mars, this Moon map was much more difficult because the geologic data was split into six different datasets. Each dataset had unique labels (and sometimes different data formats) so I spent a lot of time piecing the data together to create a cohesive map.
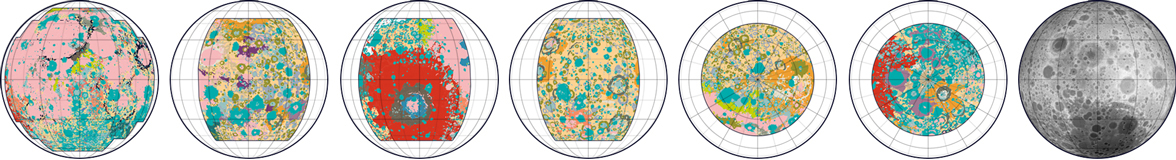
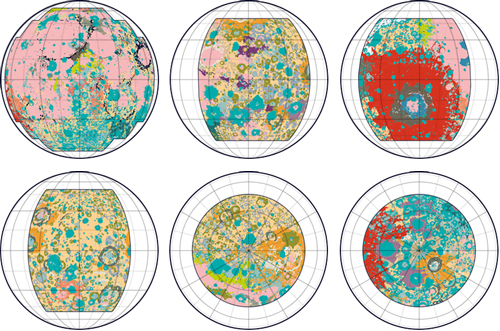
One complication with having six datasets instead of one was that some geologic categories were described differently in each dataset, like “Basin Material, Rugged” vs. “Material of Rugged Basin Terrain”. So I decided to combine closely related terms into a single color.
This Moon data was also challenging because the geologic timescales weren’t very precise. Some areas were described with uncertainty - like plains from the “Imbrian or Nectarian” era. And some geologic categories combined many time periods, like craters from “Imbrian, Nectarian, and pre-Nectarian” time periods. I thought it was too complicated to show all these uncertain aggregations in one map, so I decided to omit timescale data entirely. The map features are colored only by geologic category (craters, basins, etc.) and not by age.

 The original maps were originally published in the 70’s, and I loved the bright neon color schemes in these historical maps. I tried to keep as many of the same colors as possible, though I re-arranged to save the more extreme colors for smaller or more unusual geologic formations.
The original maps were originally published in the 70’s, and I loved the bright neon color schemes in these historical maps. I tried to keep as many of the same colors as possible, though I re-arranged to save the more extreme colors for smaller or more unusual geologic formations.
The end result is a summarized geologic map rather than a precise replication of the original data. But even though I combined geologic features and didn’t include precise scientific labels, I think the map still works in its limited role as a condensed overview of Moon geology.
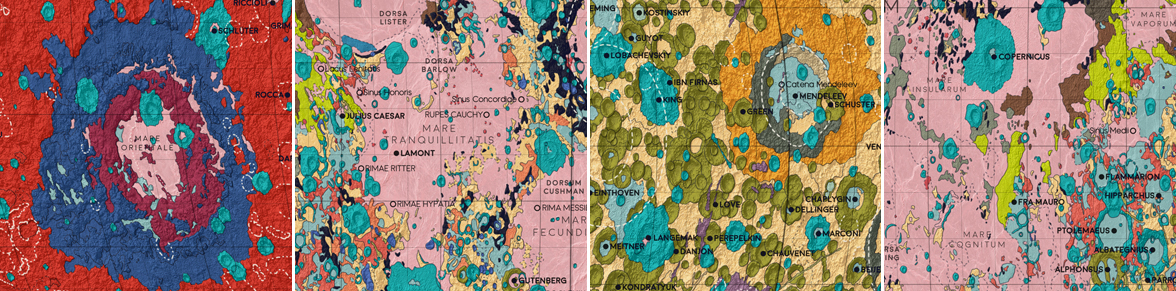
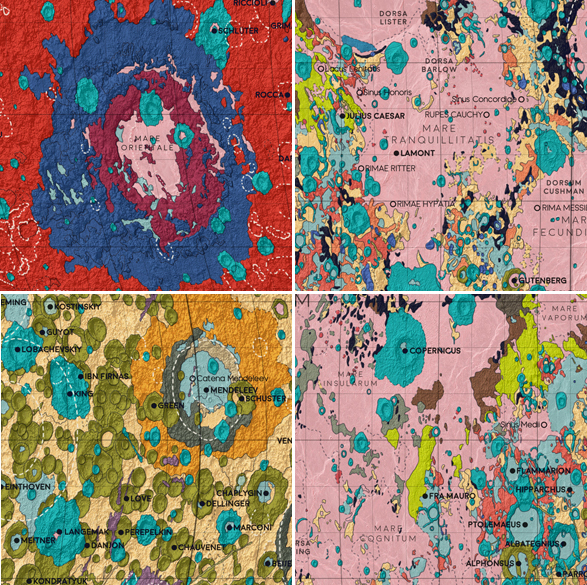 Some of the distinctive locations on the Moon. 1: Mare Orientale, a huge impact crater on the far side of the moon. 2: Mare Tranquillitatis, the landing site of Apollo 11 - the first manned landing on the Moon. 3: The far side of the moon. 4: Fra Mauro crater (at center, next to the neon green Fra Mauro highlands), the landing site of Apollo 14 in 1971.
Some of the distinctive locations on the Moon. 1: Mare Orientale, a huge impact crater on the far side of the moon. 2: Mare Tranquillitatis, the landing site of Apollo 11 - the first manned landing on the Moon. 3: The far side of the moon. 4: Fra Mauro crater (at center, next to the neon green Fra Mauro highlands), the landing site of Apollo 14 in 1971.
This map is actually the last installment in the Atlas of Space series. I love working with astronomy data and I’m sure I’ll make more maps in the future, but for now I’m taking a break to try and finish my last PhD research project. Thanks for all of your comments and suggestions this summer - I really enjoyed getting to share these maps with you! For those of you going to NACIS meeting this fall, I’m looking forward to talking more about these maps at the conference in the Using Open Data panel. Hopefully I’ll see some of you there!
-
Sources
- Data: Gazetteer of Planetary Nomenclature, International Astronomical Union (IAU). © 2019 Working Group for Planetary System Nomenclature (WGPSN). Planetary Symbology Mapping Guidelines, Federal Geographic Data Committee. LRO LOLA Elevation Model 118m (LDEM GDR). © 2018 NASA PDS and Derived Products Annex. LOLA Science Team. Lunar 5M Geologic Map Renovation. NASA PDS and Derived Products Annex. © 2016 USGS Astrogeology Science Center. Missions to the Moon. © 2019 The Planetary Society. Reference texts: Astronomy, Andrew Fraknoi, David Morrison, Sidney C. Wolff et al. © 2016 OpenStax. Fonts: The labels on this map are typeset in Moon by Jack Harvatt. The title font is RedFlower by Type & Studio. Advice: Thank you to Henrik Hargitai, Oliver Fraser, Michael Ruxton, Chloe Pursey, and Leah Willey for their helpful advice in making this map.
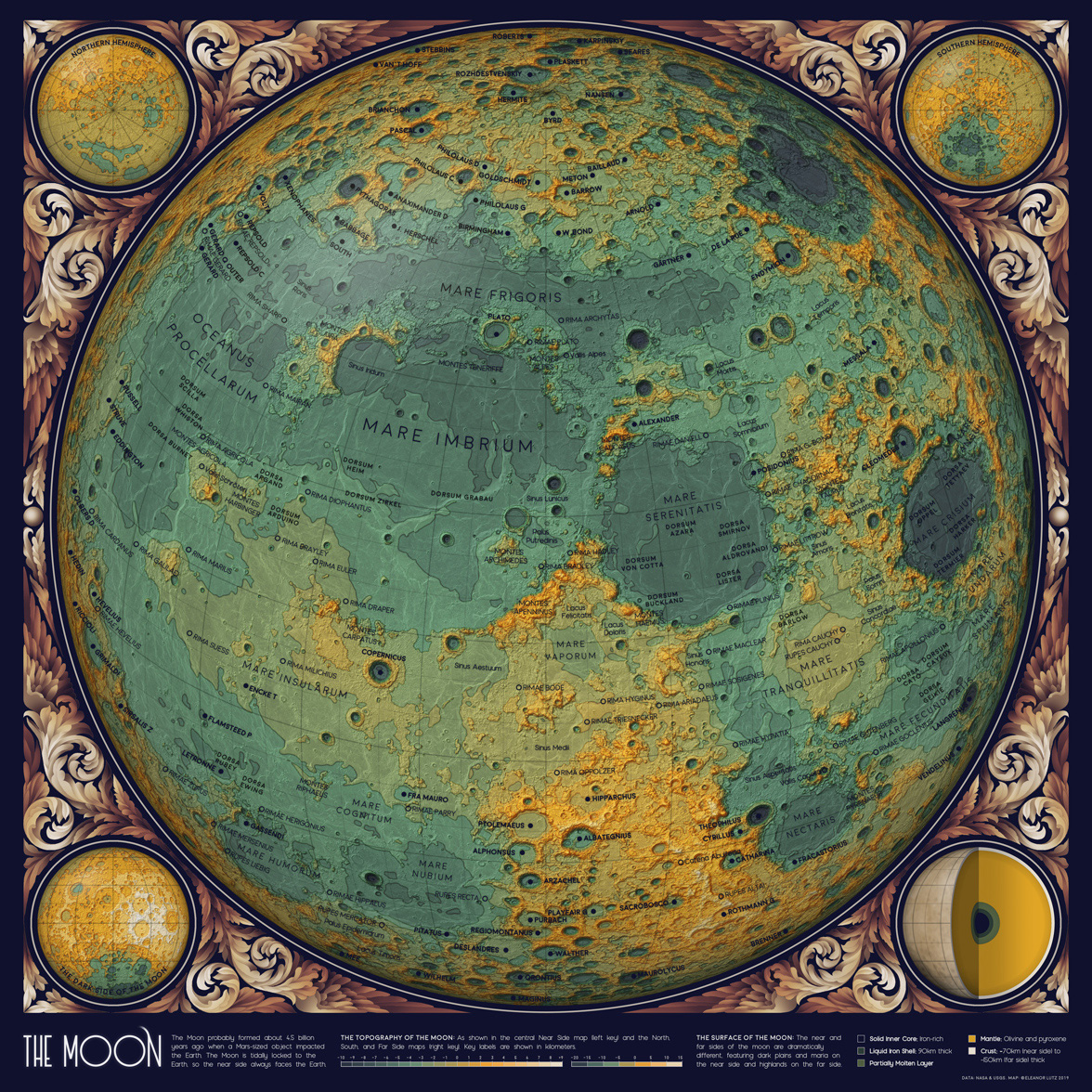
A Topographic Map of the Moon
August 19 2019 · Link to the Open-Source Code
My next two maps in the Atlas of Space series show the topography and geology of the Moon. These were some of my favorite datasets in the project - the Moon is our closest celestial neighbor, and we know a lot more about the Moon than about our neighboring planets. The Apollo missions even returned several hundred kilograms of lunar rock to the Earth for detailed analysis.
The moon is tidally locked to the Earth, which means that it completes one rotation around its axis in exactly the same time that it takes to circle around the Earth. Because of this, the far side of the moon is never visible from the Earth’s surface. This topographic map shows the near side of the moon in the large central map, as well as the North pole, South pole, far side, and interior in the four surrounding maps.
People have been fascinated by the moon for thousands of years, and lunar events like the full moon or an eclipse are tied to ancient calendars, myths, and celebrations. The lunar cycle can even be seen today in the pattern of Google searches for the word “moon,” which peaks predictably during every full moon.
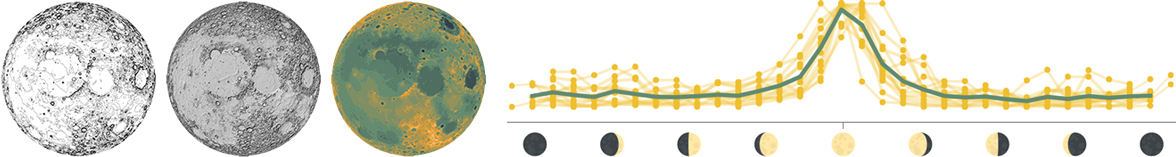
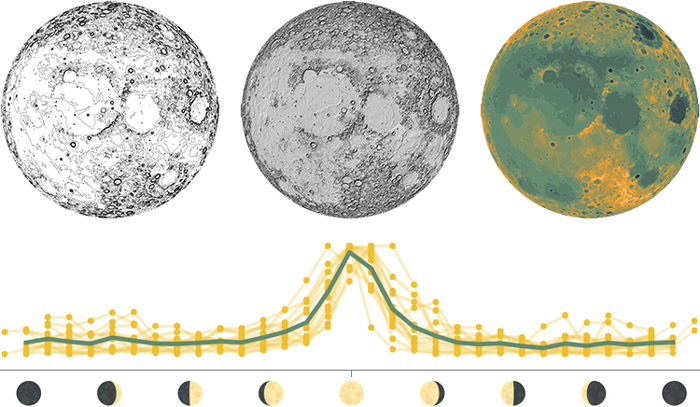 1: Data layers used to design this map. 2: Google search frequency for the word “moon” in the United States (data from Moon phase dates and Google Trends). Each yellow line shows one lunar cycle in 2018, normalized to the highest and lowest value within each lunar cycle. The turquoise line shows the overall mean.
1: Data layers used to design this map. 2: Google search frequency for the word “moon” in the United States (data from Moon phase dates and Google Trends). Each yellow line shows one lunar cycle in 2018, normalized to the highest and lowest value within each lunar cycle. The turquoise line shows the overall mean.
-
Sources
- Data: Gazetteer of Planetary Nomenclature, International Astronomical Union (IAU). © 2019 Working Group for Planetary System Nomenclature (WGPSN). Earth's Moon In Depth. © 2019 NASA Science Solar System Exploration. LRO LOLA Elevation Model 118m (LDEM GDR). © 2018 NASA PDS and Derived Products Annex. LOLA Science Team. Reference texts: Astronomy, Andrew Fraknoi, David Morrison, Sidney C. Wolff et al. © 2016 OpenStax. Fonts: The labels on this map are typeset in Moon by Jack Harvatt. The title font is RedFlower by Type & Studio. Advice: Thank you to Oliver Fraser, Henrik Hargitai, Chloe Pursey, and Leah Willey for their helpful advice in making this map.